
The influence of childhood HIV infection on the dental microbiome and tooth decay
Collaboration with Rutgers School of Dental Medicine, University of Florida, the University of Benin Teaching Hospital - Nigeria, and the Institute of Human Virology - Nigeria
Childhood caries is a serious public health problem affecting the immediate and long-term quality of life of both the child and its family, and recent studies have shown an increased prevalence of caries in HIV-infected children. Making use of a large and unique cohort of HIV+ children in Nigeria, this project will explore the influence that HIV infection has on the dental microbiome. Ultimately, the knowledge gained will translate into improved prevention and intervention strategies that could potentially mitigate an increased risk of caries in an HIV infected child.


Oral probiotics that promote dental health
Collaboration with University of Florida
The human oral biome is a diverse microbial community and previous studies have shown that some bacterial comunities may promote dental health. This project is studying the bacteria within these communities towards the development of an oral probiotic that may mitigate tooth decay.


Community dynamics of the microbiome associated with outbreaks of Legionnaires' disease
Collaboration with the Savannah River National Laboratory
Legionella are gram negative bacteria found naturally in fresh water that can cause fatal respiratory infections in humans known as Legionnaires' disease. These bacteria grow best in warm water and often colonize man-made structures such as large plumbing systems and air-conditioning cooling towers. In extensively colonized systems, the bacteria can become airborne within water vapor and cause an outbreak. Amplification of the bacterium within water systems is initiated within complex biofilms and involves a close synergy with amoeba. This project aims to better understand the community dynamics of these biofilms and the factors associated with outbreaks.
Past projects
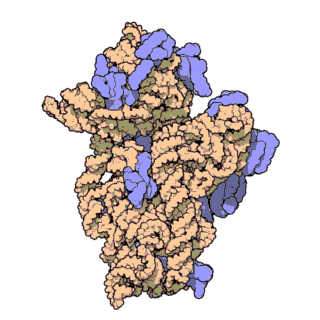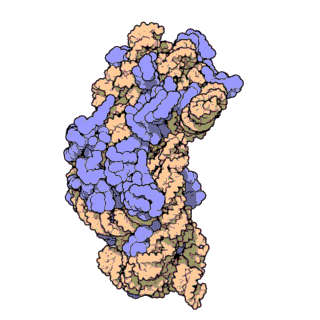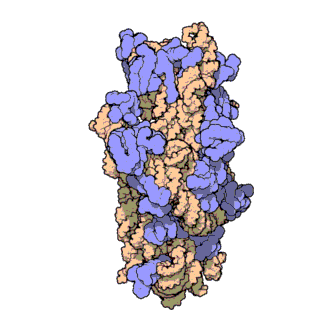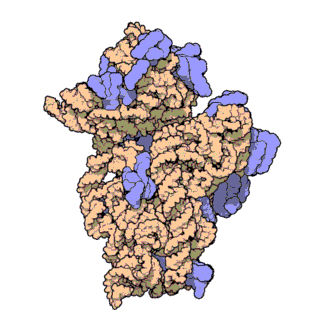
Phylogenetic signal of bacterial ribosomal genes
This study is utilizing novel phylogenomic approaches to evalute the phylogenetic signal for bacterial ribosomal genes including 16S rRNA. Our findings have important implications for microbiome studies where this gene is used extensively. Animation by David S. Goodsell



Population genomics of Streptococcus agalactiae from infected bovine and fish sources
This project is investigating genetic population structure and transmission dynamics of the zoonotic pathogen Streptococcus agalactiae. This bacteria is an important pathogen that infects multiple species including humans, livestock, and aquaculture. For example, it is (i) a leading cause of morbidity and mortality in neonates, (ii) a major cause of bovine mastitis, which is the dominant health disorder affecting milk production in the dairy industry worldwide, and (iii) an aetiological agent of septicaemia and meningo-encephalitis in saltwater and freshwater fish species. Towards a better understanding of the evolution and transmission dynamics of S. agalactiae both within and across host species, this project leverages comparative genomic and transcriptomic approaches to a global collection of hundreds of S. agalactiae strains isolated from a diverse range of host species.


Transmission dynamics of Mycobacterium avium subspecies paratuberculosis (MAP)
Collaboration with Cornell University
Mycobacterium avium subspecies paratuberculosis (MAP) is an important pathogen of cows that causes Johne's disease, with estimates suggesting that over 90% of US dairy cows are infected. Towards a better understanding of the spread of this disease, this project is utilizing population genomics to elucidate the transmission dynamics of M. avium subspecies paratuberculosis among US herds.
The Human MicroBiome



Artificial sweetners and the dental microbiome
This study is investigating how the dental microbiome responds to different artificial sweetners.
Department of Biological Sciences, College of Science, Clemson University, Clemson, SC 29634